2.1.1.356: [histone H3]-lysine27 N-trimethyltransferase
This is an abbreviated version!
For detailed information about [histone H3]-lysine27 N-trimethyltransferase, go to the full flat file.
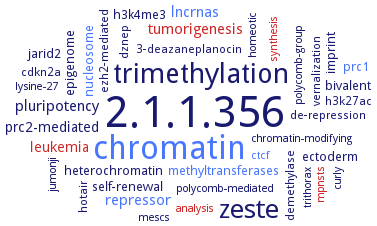
Word Map on EC 2.1.1.356 
-
2.1.1.356
-
chromatin
-
trimethylation
-
zeste
-
tumorigenesis
-
pluripotency
-
repressor
-
lncrnas
-
prc2-mediated
-
leukemia
-
heterochromatin
-
epigenome
-
ectoderm
-
methyltransferases
-
self-renewal
-
jarid2
-
bivalent
-
prc1
-
h3k4me3
-
nucleosome
-
imprint
-
cdkn2a
-
demethylase
-
dznep
-
vernalization
-
h3k27ac
-
ezh2-mediated
-
hotair
-
de-repression
-
3-deazaneplanocin
-
lysine-27
-
ctcf
-
jumonji
-
mescs
-
analysis
-
polycomb-group
-
curly
-
trithorax
-
homeotic
-
synthesis
-
mpnsts
-
polycomb-mediated
-
chromatin-modifying
- 2.1.1.356
- chromatin
-
trimethylation
-
zeste
- tumorigenesis
-
pluripotency
- repressor
- lncrnas
-
prc2-mediated
- leukemia
-
heterochromatin
-
epigenome
- ectoderm
- methyltransferases
-
self-renewal
- jarid2
-
bivalent
- prc1
-
h3k4me3
- nucleosome
-
imprint
-
cdkn2a
- demethylase
-
dznep
-
vernalization
-
h3k27ac
-
ezh2-mediated
-
hotair
-
de-repression
-
3-deazaneplanocin
-
lysine-27
- ctcf
-
jumonji
-
mescs
- analysis
-
polycomb-group
-
curly
-
trithorax
-
homeotic
- synthesis
- mpnsts
-
polycomb-mediated
-
chromatin-modifying
Reaction
3 S-adenosyl-L-methionine
+
Synonyms
CLF, EC 2.1.1.43, EZH1, EZH2, G9a, histone H3 K27 methyltransferase, histone H3 lysine 27 methyltransferase, histone lysine methyltransferase, HMTase, interleukin-5 response element II binding protein, KMT6, KMT6A, KMT6B, lysine-preferring HMTase, NSD2, NSD3, Polycomb repressive complex 2, PRC2, RE-IIBP, SET, SET domain histone lysine methyltransferase, SETDB1, TXR1, vSET, WHSC1/MMSET isoform RE-IIBP
ECTree
Advanced search results
Turnover Number
Turnover Number on EC 2.1.1.356 - [histone H3]-lysine27 N-trimethyltransferase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
0.00004
mutant enzyme Y109G, at pH 8.0 and 23°C
0.006
(+/-)-2',3'-dibenzyl-S-adenosyl-L-methionine
wild type enzyme, at pH 8.0 and 23°C
0.0217
(+/-)-2',3'-dibenzyl-S-adenosyl-L-methionine
mutant enzyme L116A, at pH 8.0 and 23°C
0.002
mutant enzyme L116A, at pH 8.0 and 23°C
0.002
(+/-)-2'-benzyl-S-adenosyl-L-methionine
wild type enzyme, at pH 8.0 and 23°C
0.000008
mutant enzyme Y109G, at pH 8.0 and 23°C
0.002
(+/-)-3'-benzyl-S-adenosyl-L-methionine
wild type enzyme, at pH 8.0 and 23°C
0.0025
(+/-)-3'-benzyl-S-adenosyl-L-methionine
mutant enzyme L116A, at pH 8.0 and 23°C
0.000008
mutant enzyme Y109G, at pH 8.0 and 23°C
0.065
S-adenosyl-L-methionine
wild type enzyme, at pH 8.0 and 23°C
0.123
S-adenosyl-L-methionine
mutant enzyme L116A, at pH 8.0 and 23°C

 results (
results ( results (
results ( top
top






